Session 2: Protein Structure Prediction Using Artificial Intelligence#
The 2024 Nobel Prize in Chemistry was awarded to Demis Hassabis and John Jumper for their groundbreaking work on AlphaFold2, an artificial intelligence system that revolutionized protein structure prediction. This session explores the journey from traditional methods to AI-powered solutions in understanding protein structures.
Understanding Proteins: Nature’s Molecular Machines#
Before we dive into prediction methods, let’s understand what proteins are and why their structure is so important.
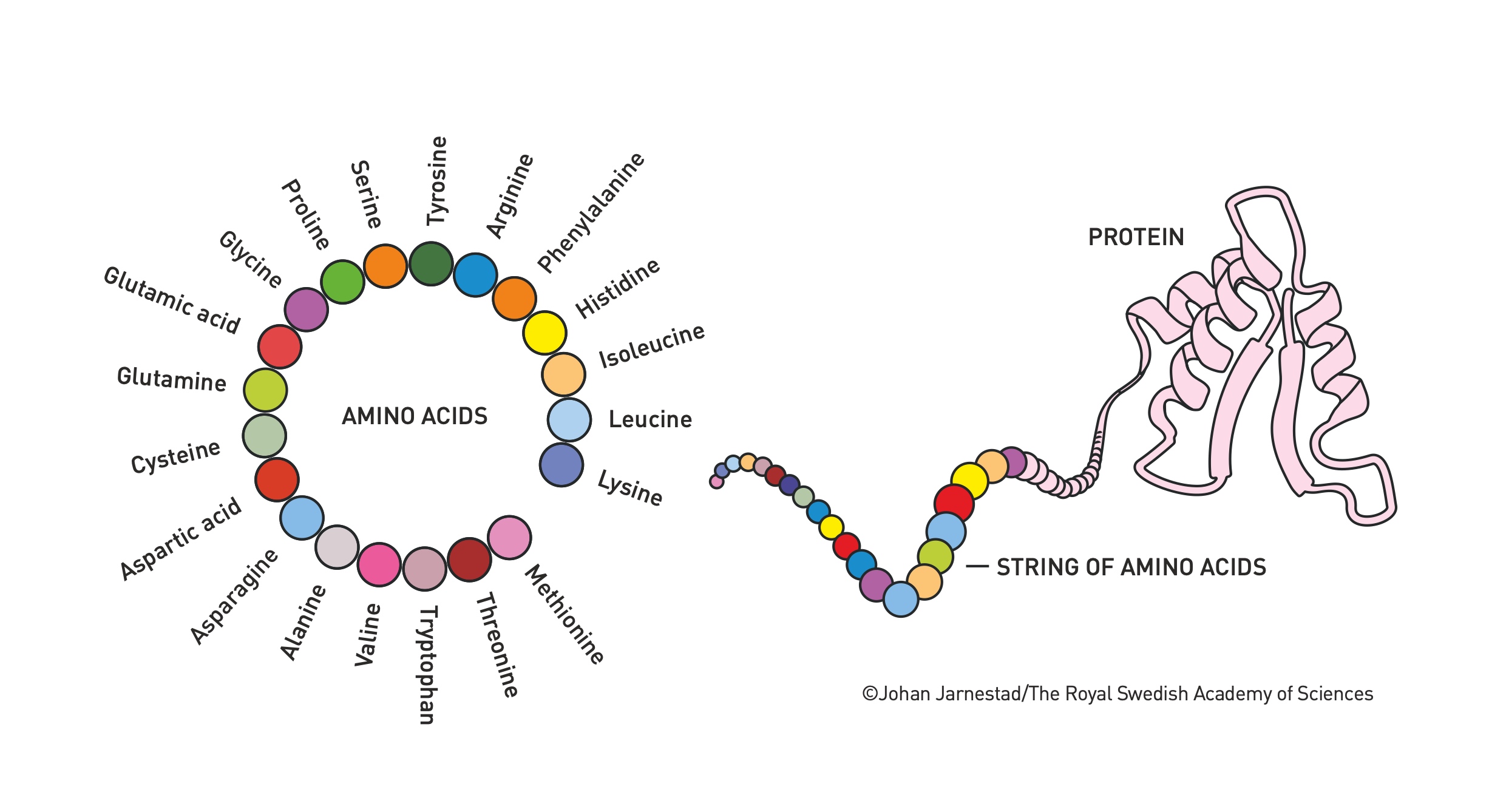
As shown in the image above, proteins are long chains of amino acids that fold into complex 3D shapes. There are 20 different types of amino acids, and their sequence determines how the protein will fold. This 3D structure is crucial because it defines the protein’s function.
Why is Protein Structure Important?#
Biological Functions: Proteins are involved in nearly every biological process, acting as:
Enzymes (catalyzing chemical reactions)
Hormones (signaling molecules)
Antibodies (defending against pathogens)
Structural components (giving cells and tissues their shape)
Drug Discovery: Understanding protein structures helps in designing drugs that can interact with specific proteins.
Disease Understanding: Many diseases occur due to misfolded proteins or mutations that affect protein structure.
The Protein Folding Challenge#
For decades, determining protein structures was a monumental task:
Experimental Methods:
X-ray crystallography and NMR spectroscopy were the primary methods.
These methods are time-consuming, expensive, and don’t work for all proteins.
The Prediction Problem:
Scientists have long sought to predict structure from amino acid sequence alone.
This is incredibly difficult due to the astronomical number of possible configurations (known as Levinthal’s paradox).
CASP Competition:
Started in 1994, the Critical Assessment of Protein Structure Prediction (CASP) aimed to track progress in this field.
For years, improvements were slow and incremental.
Enter AlphaFold: AI Tackles Protein Folding#
In 2018, DeepMind, led by Demis Hassabis, entered the CASP competition with AlphaFold, marking the beginning of a revolution.
AlphaFold2: A Quantum Leap#
In 2020, AlphaFold2 achieved near-experimental accuracy in protein structure prediction, solving a 50-year-old grand challenge in biology.
How Does AlphaFold2 Work?#
Let’s break down the process:
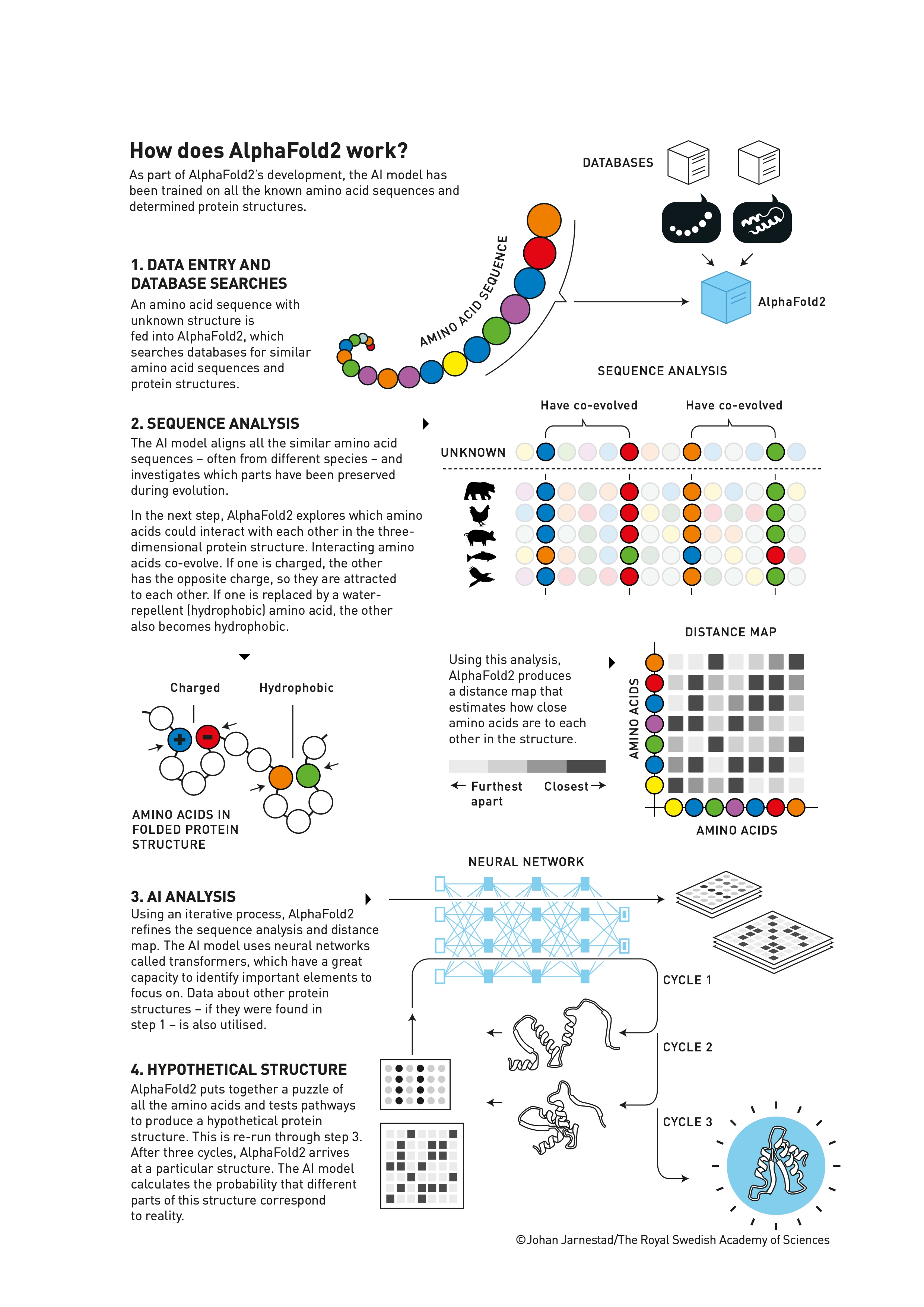
Data Input:
The system takes an amino acid sequence as input.
Database Search:
It searches for similar sequences in protein databases.
Evolutionary Analysis:
AlphaFold2 aligns similar sequences to identify regions that have evolved together, providing clues about the structure.
Distance Mapping:
The AI creates a “distance map” predicting how close different amino acids will be in the final 3D structure.
Transformer Neural Networks:
These are the secret sauce of AlphaFold2. Transformers can process entire sequences at once, finding complex patterns and relationships.
They’re excellent at understanding context, which is crucial for predicting how distant parts of a protein might interact.
Iterative Refinement:
The system goes through multiple cycles, refining its prediction each time.
Final Structure:
The output is a highly accurate 3D model of the protein.
The Impact of AlphaFold2#
The implications of this breakthrough are vast:
Speed: What once took years can now be done in minutes.
Accessibility: The code is open-source, democratizing protein research.
Wide Adoption: By 2024, over two million researchers worldwide were using AlphaFold2.
Real-World Applications#
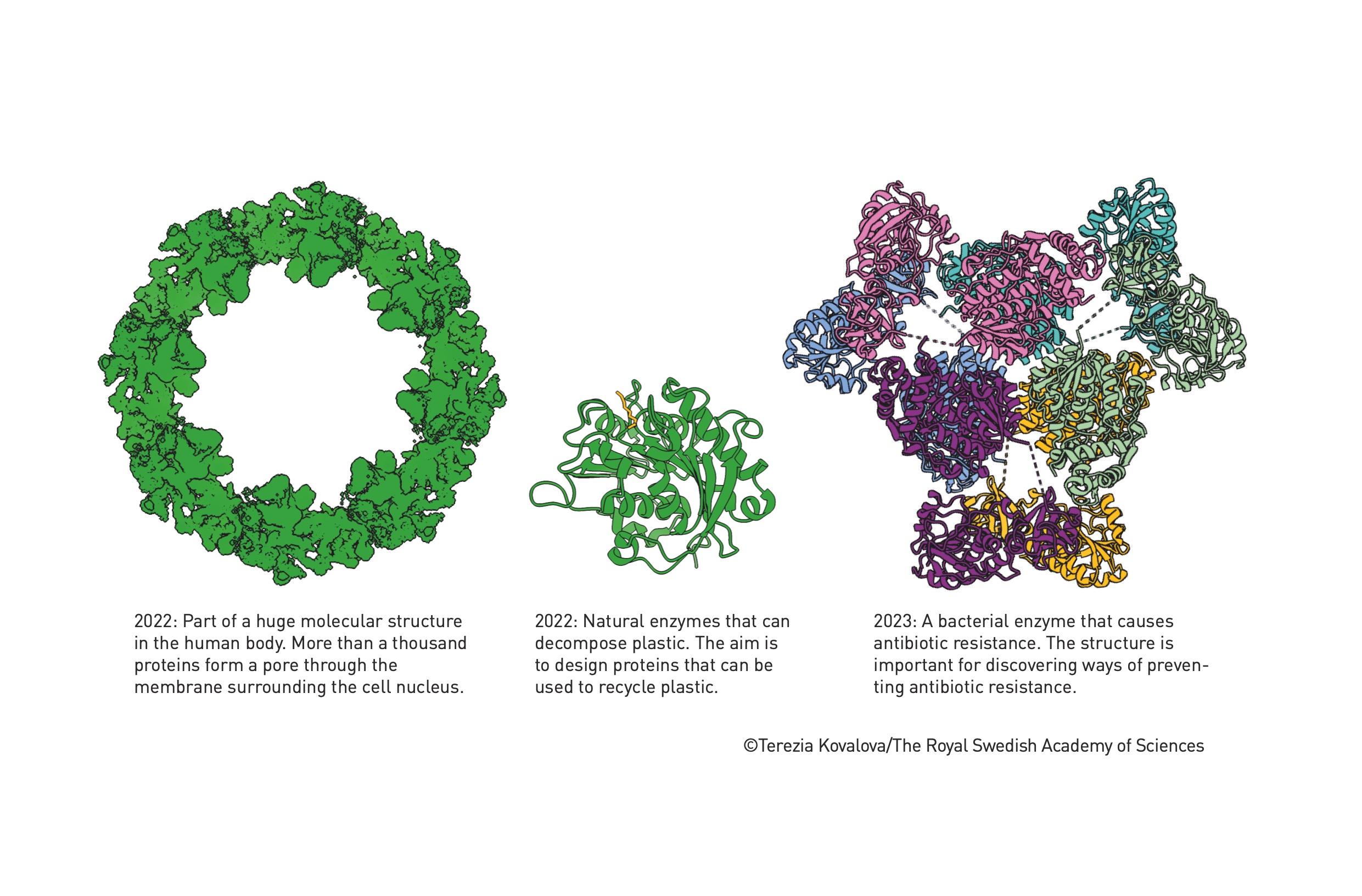
Drug Development:
Faster identification of drug targets.
Better understanding of how drugs interact with proteins.
Environmental Solutions:
As shown in the image, researchers are using AlphaFold2 to study enzymes that can break down plastics, potentially revolutionizing recycling.
Disease Research:
Understanding protein misfolding in diseases like Alzheimer’s.
Synthetic Biology:
Designing new proteins for specific functions.
The Future of Protein Science#
AlphaFold2 has opened new horizons:
Integration with Other Methods: Combining AI predictions with experimental techniques for even better results.
Protein Design: Moving from prediction to designing entirely new proteins with desired functions.
Systems Biology: Understanding how proteins interact in complex biological systems.
Key Takeaways#
AI Revolution: Deep learning and transformer models have solved a decades-old problem in biochemistry.
Democratization of Science: Open-source AI tools are accelerating research across various fields.
Interdisciplinary Impact: From medicine to environmental science, the applications are vast and growing.
Ongoing Challenge: While structure prediction is largely solved, understanding protein dynamics and interactions remains a frontier.
The work of Hassabis, Jumper, and the DeepMind team has not just advanced our understanding of proteins; it has fundamentally changed how we approach complex biological problems. As we continue to explore the potential of AI in science, we can expect even more groundbreaking discoveries at the intersection of technology and biology.